Emerging infectious diseases such as Zika, Ebola, Chikungunya and MERS are dangerous and unpredictable. Public health
organizations need data as early as possible to predict disease spread and plan responses. Yet early data is very
difficult to obtain, because it must be proactively collected from potential disease sources in the environment.
Project Premonition aims to detect pathogens before they cause outbreaks — by turning mosquitoes into devices that collect
data from animals in the environment. There are over 3,600 known species of mosquitoes, which bite a wide range of animals
from dogs and chickens to snakes and mice. Each bite may collect a few microliters of blood, containing genetic information
about the animal that was bitten and pathogens circulating in that animal. In fact, it has already been shown that the DNA
collected from mosquitoes can be used to identify: (1) the types of animals that were bitten, (2) mosquito-borne diseases
such as Zika and West Nile that infect both mosquitoes and hosts (e.g. humans and animals), and (3) previously unknown viruses of unknown origin.
The complete set of genes or genetic material present in a cell or organism.
A DNA sample containing the genomes of multiple organisms.
The process of identifyting the individual genomes represented in a metagenome. This process is akin to dumping hundreds of jigsaw puzzles onto a table and trying to solve them all at once.
Known genomes previously isolated and sequenced to be used for assembly. They are stored in a digital library.
The process of arranging the sequences of DNA to identify regions of similarity between two sequences.
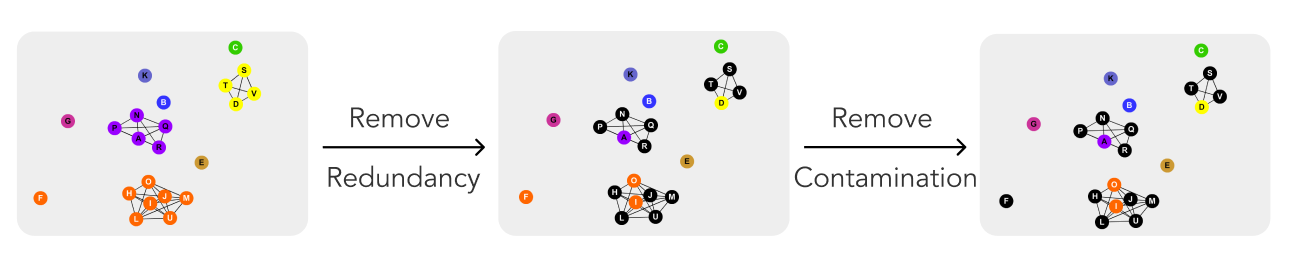
The precise goal is to optimize the pool of reference genomes.
First, we will remove redundant genomes (genomes that have another perfect match) from the pool of reference genomes.
- Break each file down into pieces.
- Group files by size (only files of the same size can be a perfect match).
- Check if these grouped genomes match any others using alignment.
- Keep only unique genomes. Toss/Flag replicates.
Next, we will remove contaminated genome samples (genome samples that have more than one genome in them).
- Align each viral and bacterial genome against every other sample.
- Flag any genome with a match to get a baseline.
- Group flagged genomes by phylogeny.
- Flag genomes that are significantly different from their closest relatives. If there are no close relatives, the genome will remain flagged, but will not be removed.

